Review of HTA Frameworks and Decisions for Next-Generation Sequencing in Precision Oncology
By Xcenda
HTA QUARTERLY | SUMMER 2019
Review of HTA Frameworks and Decisions for Next-Generation Sequencing in Precision Oncology
Date last updated: August 8, 2019
Precision Medicine in Oncology

Advances in Genomic Testing: NGS
Prior to the advent of NGS, DNA sequencing was labor intensive and costly. However, second-generation sequencing technologies, commonly referred to as NGS, have been developed in an effort to enable the routine genomic study of every tumor. NGS allows for rapid and accurate sequencing of many genes at once utilizing either DNA or RNA. For most clinical applications, NGS uses gene panels to sequence only a discrete number of genes of interest, making it less labor intensive than complete DNA or RNA sequencing methods.
The benefit of NGS technology is that it saves costs compared to multiple, individual nucleic acid-based tests (such as fluorescent in situ hybridization [FISH], polymerase chain reaction [PCR], etc). The small amount of tumor tissue required may also eliminate the need for an additional procedure, such as a repeat biopsy. However, there are limitations associated with NGS technology, including technical complexity and cost in comparison to other testing methodologies, and reimbursement is often restricted.
Outcomes Evidence Supporting Precision Medicine in Oncology
Precision medicine aims to find patients with targetable oncogenic drivers to provide a therapeutic intervention expected to have greater clinical benefit based on the specific molecular or cellular features of the tumor. Recent studies report precision medicine improves outcomes in patients with actionable alterations for which there is targeted therapy available compared with standard of care chemotherapy or best supportive care. Further, studies have shown as many as 30% to 40% of patients with advanced cancer who undergo tumor genomic profiling have an actionable alteration that can be matched to a Food and Drug Administration (FDA)-approved targeted therapy. Although this advance in technology is leading to novel therapeutic strategies, one question continually being asked is, “Does an ‘actionable alteration’ always yield an important gain in improved outcomes at an acceptable increase in costs?”
Targeted therapy has moved more into the spotlight, as the advantages associated with NGS over other detection technologies have the potential to lift molecular diagnostics in clinical oncology to the next level. However, there is currently a lack of direct evidence linking NGS to improved clinical outcomes in oncology (eg, increased survival). This lack of direct evidence for NGS is a 2-fold problem. First, efficacy and safety outcomes data, including survival endpoints, from randomized controlled trials for precision medicine are not yet mature. Second, since many of these types of studies do not separate out based on methodology used to determine the genetic alteration, there is a need for assessment of individual methodologies in terms of both efficacy and cost.
As NGS technology evolves and becomes further incorporated into routine clinical practice, the need for data on NGS in oncology is becoming evident. One economic study showcased at the American Society of Clinical Oncology Annual Meeting in 2018 assessed the economic impact of NGS vs sequential single-gene testing modalities in patients with metastatic non-small cell lung cancer (NSCLC) in the United States (US). The results of this analysis supported upfront NGS use to establish genomic alteration status for newly diagnosed patients with metastatic NSCLC to inform treatment decisions; the wait times for results were generally shorter, and payer cost was lowest with NGS.
HTA Frameworks and Decisions for NGS
Health technology assessment (HTA) agencies’ evaluations with precision medicine have so far primarily examined diagnostic and companion diagnostic tests. Several countries have accommodated the additional complexities of evaluating precision medicine tests through new procedures, such as the Diagnostic Assessment Programme at the National Institute for Health and Care Excellence (NICE) in England or the Health Technology Assessment Access Point in Australia. However, the majority of HTAs and payers have not tackled the tough topic of NGS with definitive guidance on use of the technology.
Several decisions have forced at least partial opinions on NGS to begin to shape the coverage landscape and attitude toward NGS, as shown in the examples below.
- In 2014, the Canadian Agency for Drugs and Technologies in Health (CADTH) conducted a review of cost-effectiveness and guidelines associated with NGS DNA sequencing technology. CADTH concluded that there is limited evidence to establish the cost-effectiveness of NGS technology and that there were no established standardized guidelines identified. The guidelines that were identified only described high-level recommendations on implementation of the technology, but recommendations for specific applications were lacking. CADTH further concluded that as additional advances are made in NGS development and data analysis, robust protocols will need to be developed to establish clinical methodologies regarding the cost-effectiveness of this technology in clinical practice.
- NICE has not yet provided overall guidance concerning NGS technology use in oncology broadly, though individual assessments have been made within particular disease states. Take, for example, diagnostic guidance for epidermal growth factor receptor (EGFR) mutation testing in adults with locally advanced or metastatic NSCLC. When this guidance was published in 2013, there was insufficient evidence to make any recommendation on the use of NGS testing for EGFR mutational analysis per the NICE assessment.
- In 2018, the US Centers for Medicare & Medicaid Services (CMS) made a coverage determination for NGS technology, stating that it would cover NGS testing for patients with recurrent, relapsed, refractory, metastatic cancer. Additionally, patients with advanced stages III or IV cancer would be covered if the test is FDA approved as a companion diagnostic for that patient’s cancer type only if the NGS test has not previously been used. However, repeat testing would be approved in patients with a new primary cancer diagnosis or in those patients deciding to seek further treatment.
- Medicare Administrative Contractors (MACs) may determine coverage of other non-FDA-approved companion diagnostic laboratory tests using NGS for patients with cancer only if the above criteria are met.
- Medicare Administrative Contractors (MACs) may determine coverage of other non-FDA-approved companion diagnostic laboratory tests using NGS for patients with cancer only if the above criteria are met.
Ongoing Precision Medicine and NGS Research
Given the lack of evidence around cost-effectiveness and clinical use, it is increasingly important that future studies track efficacy of these targeted therapies. Measures such as overall survival (OS), progression-free survival (PFS), overall response rate (ORR), and other data will be useful for evaluating sequencing-matched therapies’ effect on patient outcomes to inform future treatment and insurance coverage standards.
Although the data are currently limited, there are several ongoing studies, highlighted below, that may provide more clarity regarding the efficacy of precision medicine, including NGS technology, for oncology patients. These studies aim to learn from the real-world practice of prescribing targeted therapies to patients with advanced cancer whose tumor harbors a genomic variant known to be a drug target or to predict sensitivity to a drug.
Table 1. Clinical Trials Utilizing Precision Medicine-Based Treatment Approach in Oncology
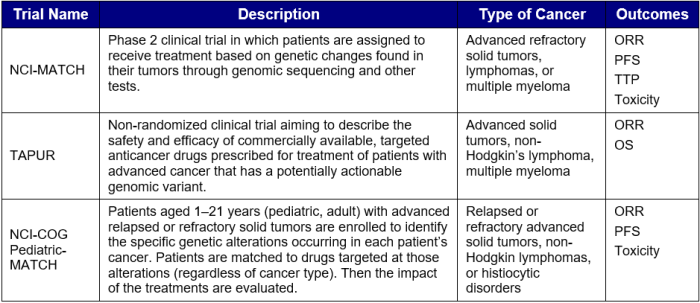
Key: ORR – overall response rate; OS – overall survival; PFS – progression-free survival; TTP – time to progression.
Cost Considerations for NGS
With more efficacy data associated with precision medicine on the horizon, the need for cost data associated with NGS becomes increasingly evident. As there are a multitude of different types of testing technologies available to determine a patient’s potential ability to benefit from targeted therapy, HTA agencies, payers, and policymakers will need to be informed on the most cost-effective strategy to align patients with the correct therapy. As a result, some key questions surrounding costs that stakeholders will need to address with regard to NGS include:
- How will utilizing NGS affect total healthcare costs?
- What additional data will manufacturers need to produce to support the pricing of targeted therapies resulting from NGS use?
- What type of cost-effectiveness measure, cost per life-year gained, or cost per quality-adjusted life-year (QALY)—or a different measure—should be used to capture the value of NGS?
- How does the cost-effectiveness and value of NGS compare to using a single mutation test?
Gaps in the Current HTA Reviews of NGS
NGS as a diagnostic, prognostic, and therapeutic guidance technology has not been assessed by most HTA agencies to date. The HTAs agencies that have assessed NGS have concluded that there is not enough evidence to support its cost-effectiveness or use in clinical practice. Although CMS has made a coverage determination for NGS, both patients and the NGS test used must meet strict requirements for coverage. Therefore, there is a distinct need for both data supporting NGS technology as well as guidelines for implementation and appropriate use of such technology in clinical practice to guide HTA agencies and policy decision makers in the future.
Conclusion
As the realm of precision medicine within oncology continues to expand, the need for information surrounding the appropriate use and value of genomic alteration testing technology, including NGS, becomes more apparent. While few HTA agencies have addressed NGS technology, this is likely to change as more efficacy data associated with precision medicine become available. What remains to be seen is how this information will help in distinguishing between various types of testing in order for HTA agencies and payers to make decisions regarding the use of various testing strategies within oncology clinical practice. There is a distinct need for comparative data analyzing the cost-effectiveness of the various testing strategies to inform policymaking regarding NGS testing.
For further information about this topic, please also see the article on tumor-agnostic therapy assessment in this issue of HTA Quarterly.
The article should be referenced as follows:
Pollack M, Sorenson F, Redekop K, Campbell C. Review of HTA Frameworks and Decisions for Next-Generation Sequencing in Precision Oncology. HTA Quarterly. Summer 2019. August 27, 2019.
Sources
- Alekseyev YO, Fazeli R, Yang S, et al. A next-generation sequencing primer—how does it work and what can it do? Acad Pathol. 2018 May 6;5:2374289518766521.
- Boland GM, Piha-Paul SA, Subbiah V, et al. Clinical next generation sequencing to identify actionable aberrations in a phase I program. Oncotarget. 2015;6(24):20099-20110.
- CADTH. Next generation DNA sequencing: a review of the cost effectiveness and guidelines. February 6, 2014. https://cadth.ca/next-generation-dna-sequencing-review-cost-effectiveness-and-guidelines. Accessed May 13, 2019.
- ClinicalTrials.gov. Targeted Therapy Directed by Genetic Testing in Treating Patients With Advanced Refractory Solid Tumors, Lymphomas, or Multiple Myeloma (The MATCH Screening Trial). May 23, 2019. https://clinicaltrials.gov/ct2/show/NCT02465060. Accessed May 24, 2019.
- ClinicalTrials.gov. Targeted Therapy Directed by Genetic Testing in Treating Pediatric Patients With Relapsed or Refractory Advanced Solid Tumors, Non-Hodgkin Lymphomas, or Histiocytic Disorders (The Pediatric MATCH Screening Trial). May 17, 2019. https://clinicaltrials.gov/ct2/show/record/NCT03155620?term=NCI-COG+pediatric+MATCH&rank=1. Accessed May 24, 2019.
- ClinicalTrials.gov. TAPUR: Testing the Use of Food and Drug Administration (FDA) Approved Drugs That Target a Specific Abnormality in a Tumor Gene in People With Advanced Stage Cancer (TAPUR). February 25, 2019. https://clinicaltrials.gov/ct2/show/NCT02693535. Accessed May 24, 2019.
- CMS. National Coverage Determination (NCD90.2): Next Generation Sequencing (NGS). March 16, 2018. https://www.cms.gov/Outreach-and-Education/Medicare-Learning-Network-MLN/MLNMattersArticles/Downloads/MM10878.pdf. Accessed May 13, 2019.
- Collins, FS. Shattuck lecture—medical and societal consequences of the Human Genome Project. N Engl J Med. 1999;341:28-37.
- Genetics Home Reference. National Institutes of Health. April 3, 2018. https://ghr.nlm.nih.gov/. Accessed April 13, 2018.
- Harris MH, DuBois SG, Glade Bender JL, et al. Multicenter feasibility study of tumor molecular profiling to inform therapeutic decisions in advanced pediatric solid tumors: the individualized cancer therapy (iCat) study. JAMA Oncol. 2016;2(5):608-615.
- Haslem DS, Van Norman SB, Fulde G, et al. A retrospective analysis of precision medicine outcomes in patients with advanced cancer reveals improved progression-free survival without increased health care costs. J Oncol Pract. 2017 Feb;13(2):e108-e119.
- Horak P, Fröhling S, Glimm H. Integrating next-generation sequencing into clinical oncology: strategies, promises and pitfalls. ESMO Open. 2016 Nov 18;1(5):e000094.
- Hyman DM, Taylor BS, Baselga J. Implementing genome-driven oncology. Cell. 2017 Feb 9;168(4):584-599.
- Jarvis LM. Cancer, redefined. Chemical & Engineering News. 2017;95(27):26-30.
- Joosten SEP, Retèl VP, Coupé VMH, et al. Scenario drafting for early technology assessment of next generation sequencing in clinical oncology. BMC Cancer. 2016 Feb 6;16:66.
- Kamps R, Brandão RD, Bosch BJ, et al. Next-generation sequencing in oncology: genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci. 2017 Jan 31;18(2).
- Love-Koh J, Peel A, Rejon-Parrilla JC, et al. The future of precision medicine: potential impacts for health technology assessment. Pharmacoeconomics. 2018 Dec;36(12):1439-1451.
- Marquart J, Chen EY, Prasad V. Estimation of the percentage of US patients with cancer who benefit from genome-driven oncology. JAMA Oncol. 2018 Aug 1;4(8):1093-1098.
- Massard C, Michiels S, Ferte C, et al. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov. 2017;7(6):586-595.
- Morash M, Mitchell H, Beltran H, et al. The role of next-generation sequencing in precision medicine: a review of outcomes in oncology. J Pers Med. 2018 Sep 17;8(3).
- Muzzey D, Evans EA, Lieber C. Understanding the basics of NGS: from mechanism to variant calling. Curr Genet Med Rep. 2015;3(4):158-165.
- NICE. EGFR‑TK mutation testing in adults with locally advanced or metastatic non-small-cell lung cancer. August 2013. https://www.nice.org.uk/guidance/dg9/chapter/1-Recommendations. Accessed May 13, 2019.
- Parsons DW, Roy A, Yang Y, et al. Diagnostic yield of clinical tumor and germline whole-exome sequencing for children with solid tumors. JAMA Oncol. 2016;2(5):616-624.
- Pennell NA, Mutebi A, Zhou ZY, et al. Economic impact of next generation sequencing vs sequential single-gene testing modalities to detect genomic alterations in metastatic non-small cell lung cancer using a decision analytic model. J Clin Oncol. 2018;36(15):9031-9031.
- Schwaederle MC, Zhao MM, Lee JJ, et al. Impact of precision medicine in refractory malignancies: a meta-analysis of 13,203 patients in phase I clinical trials. J Clin Oncol. 2016;34(15_suppl):11520.
- Shabani Azim F, Houri H, Ghalavand Z, Nikmanesh B. Next generation sequencing in clinical oncology: applications, challenges and promises: a review article. Iran J Public Health. 2018 Oct;47(10):1453-1457.
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013 Mar 29;339(6127):1546-1558.
- Yates LR, Seoane J, Le Tourneau C, et al. The European Society for Medical Oncology (ESMO) Precision Medicine Glossary. Ann Oncol. 2018 Jan 1;29(1):30-35.


